An Interest In:
Web News this Week
- April 14, 2024
- April 13, 2024
- April 12, 2024
- April 11, 2024
- April 10, 2024
- April 9, 2024
- April 8, 2024
Analysis of ECS 236th meeting abstracts(1) - data scraping with BeautifulSoup4
Introduction
Recently, I've published an article about introduction of my webapp, ECS Meeting Explorer.
ECS Meeting Explorer - Explore ECS meeting easily and deeply
I will give an explanation of key technique used in this app.
Main topic of this article is data scraping from .html file with BeautifulSoup4.
Environment
In the series of article, I will use Python. Please install these libraries.
- numpy > 1.14.5
- pandas > 0.23.1
- matplotlib > 2.2.2
- beautifulsoup4 > 4.6.0
- gensim > 3.4.0
- scikit-learn > 0.19.1
- scipy > 1.1.0
Jupyter notebook is also available on my GitHub repository. Check it!
Download meeting abstract
First of all, we need to get whole data of meeting abstract.
As I mentioned in before article, we can download it as .zip from below.
https://www.electrochem.org/236/download-abstracts
After unzipped, please place it in same directory as jupyter-notebook.
Please keep in mind that it will expire in 9/30/2020.
Data Scraping from table of contents
As I mentioned, We use beautifulsoup4 to scraping data.
In the ECS meeting, all presentations are categorized as several research fields.
# Load Table of Contentssoup = bs4.BeautifulSoup(open('./ECS MA2019-02.003/Abstracts/tos.html', encoding='utf-8'),'lxml')# Function to remove html tagspx = re.compile(r"<[^>]*?>")#Load Categoriesp_len = len(soup('h2'))categorys = []for i in range(p_len): c = px.sub("", str(soup('h2')[i])) categorys.append(c)print("categoly_num ="+str(p_len))print(categorys)This is a result,
- 'ABatteries and Energy Storage'
- 'BCarbon Nanostructures and Devices'
- 'CCorrosion Science and Technology'
- 'DDielectric Science and Materials'
- 'EElectrochemical/Electroless Deposition'
- 'FElectrochemical Engineering'
- 'GElectronic Materials and Processing'
- 'HElectronic and Photnonic Devices and Systems'
- 'IFuel Cells, Electrolyzers, and Energy Conversion'
- 'JLuminescence and Display Materials, Devices, and Processing'
- 'KOrganic and Bioelectrochemistry'
- 'LPhysical and Analytical Electrochemistry
- Electrocatalysis, and Photoelectrochemistry'
- 'MSensors'
- 'ZGeneral Topics'
There are 14 major categories in ECS 236th meeting.
Next, there are sub-categories for each. These are scraped from toc.html as follows,
#Load Sessionsp_len = len(soup('h3'))sessions = []for i in range(p_len): c = px.sub("", str(soup('h3')[i])) sessions.append(c)print("session_num ="+str(p_len))print(sessions)There are 58 sub-categories. Too much to write it down!
Scrape abstract texts and properties of presentation
Now, let's begin to scrape abstract texts of presentation.
Before that, please load a stop word list.
#Load stopwordswith open('stopwords.txt', 'r', encoding='utf-8') as f: stops = f.readlines()stopwords = []for s in stops: s = s.replace("\n", "") stopwords.append(s)This is a list of words filtered out before processing, such as pronoun, ordinary verbs and so on. It is necessary for improving word embedding and building a document vectors (will be explained in another article).
You can find stop word list as .txt format on a GitHub repository.
If you clone my GitHub repositry, it already contains a stopwords.txt.
Code for meeting abstract scraping as follows,
dic_all = []# make session number and name dictionarydic_sessions = {}for s in sessions: id_ = s.split('-',1) dic_sessions[id_[0]]=id_[1]#Get all pathes of abstract .html filepath = os.listdir("./ECS MA2019-02.003/Abstracts/Html")#Scraping html documentsfor n,i in enumerate(tqdm(path)): soup = bs4.BeautifulSoup(open("./ECS MA2019-02.003/Abstracts/Html/"+i, encoding='utf-8'), "html5lib") #Get title, authors, affiliation title =px.sub("", soup.title.string) author = px.sub("", str(soup('p')[1])) author = author.split(', ') affiliation = px.sub("", str(soup('p')[2])) affiliation = affiliation.replace("a", "", 1) #Get presentation number, session number = re.search('-[0-9]{4}', str(title)) number = number.group().replace("-", "") session = re.match('.*[0-9]{2}', str(title)) session = session.group() #Remove session name in title title = re.sub('\w[0-9]{2}-[0-9]{4}', "", title) title = title.replace(u"\xa0",u"") title = title.replace(session, '') title = title.replace('(Invited)', '') session = session.replace(number, '') session = session.replace('-', '') try: session_name = dic_sessions[session] except: session = session.split(u'\xa0',1)[0] session_name = dic_sessions[session] session_name = session_name.replace(u'&', '&') #Get main text and reshape it content = px.sub("", str(soup.find(class_=re.compile('contaner_content')))) stoplist = [",", ".", ":", ";", "(", ")", "{", "}", "[", "]", "\'", "\""] for s in stoplist: content = content.replace(s, "") content = content.replace("\t", "") #Get word list for Word2Vec Learning c_list = content.split(" ") c_list = [c.lower() for c in c_list] for s in stopwords: c_list = [c for c in c_list if i != s] c_list = [c for c in c_list if c != ""] #Summarize it as dictionary dic ={'num':number,'title':title,'author':author, 'affiliation':affiliation,'session':session, 'session_name':session_name, 'contents':content, 'mod_contents': c_list, 'vector':n, 'url':i} dic_all.append(dic)I'm sorry for these unsophisticated codes, but it works, anyway.
The details of all presentation including abstract text are contained as dictionary format, this is an example,
{'num': '0001', 'title': 'The Impact of Coal Mineral Matter (alumina and silica) on Carbon Electrooxidation in the Direct Carbon Fuel Cell', 'author': ['Simin Moradmanda', 'Jessica A Allena', 'Scott W Donnea'], 'affiliation': 'University of Newcastle', 'session': 'A01', 'session_name': 'Battery and Energy Technology Joint General Session', 'contents': 'Direct carbon fuel cell DCFC as an electrochemical device...', 'mod_contents': ['direct','carbon','fuel','cell', ... ,'melting'], 'vector': 0, 'url': '1.html'}'contents' and 'mod_text' are abstract text without/with preprocessing.
Data analysis of presentation in ECS meeting
We got ECS 236th meeting abstracts as a format easy to analyze.
Then, before language analysis, how about a statistical analysis?
Now, Lets calculate three metrics, number of presentation, number of authors, and number of affiliations for each 14 categories.
charactors = [i[0] for i in categorys]cat_counts = np.zeros(len(charactors))authors_counts = {}words_counts = {}for c in charactors: authors_counts[c] = [] words_counts[c] = []for d in dic_all: char = d['session'][0] cat_counts[charactors.index(char)] += 1 authors_counts[char].append(len(d['author'])) words_counts[char].append(len(d['contents'].split(' ')))Firstly, I will show the number of presentation of each categories.
fig = plt.figure(figsize=(10,4),dpi=200)plt.bar(charactors, cat_counts, label=categorys)plt.xlabel('Categories',fontsize=14)plt.title('Number of presentation',fontsize=16)plt.show()This plot shows that presentations are not distributed equally.
The most popular category is A - 'Batteries and Energy Storage', 711 counts. The second one is I - 'Fuel Cells, Electrolyzers, and Energy Conversion', 655 counts. The counts of these two categories reach more than half of total amounts(2461) of presentations.
This fact is reasonable because these two topics are related to the energy problem. It means many budgets injected to this field.
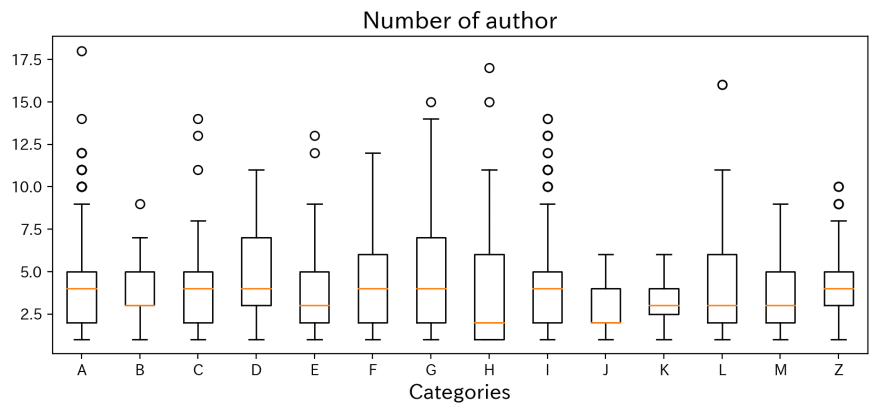
points = tuple(authors_counts[i] for i in charactors)fig = plt.figure(figsize=(10,4),dpi=200)plt.boxplot(points)plt.gca().set_xticklabels(charactors)plt.xlabel('Categories',fontsize=14)plt.title('Number of author',fontsize=16)plt.show()This is a boxplot shows how many authors named on. Actially, it seems no trends exist for each categories.
The presentation with the largest number of authors is No. 0701, 'Comparison of Correlated Li+ Diffusion from Molecular Dynamics of Promising Solid Electrolytes' listed 18 member from 9 different affiliation!
Finally, I will show the top 20 list of author's affiliation.
import collectionsaffiliation_list = []for a in dic_all: affiliation_list.extend(a['affiliation']) a_count = collections.Counter(affiliation_list)a_sorted = sorted(a_count.items(), key=lambda x:x[1], reverse=True)| Rank | Affiliation | Counts |
|---|---|---|
| 1 | Georgia Institute of Technology | 103 |
| 2 | Argonne National Laboratory | 73 |
| 3 | National Renewable Energy Laboratory | 56 |
| 4 | Lawrence Berkeley National Laboratory | 54 |
| 5 | Oak Ridge National Laboratory | 48 |
| 6 | University of South Carolina | 41 |
| 7 | Sandia National Laboratories | 33 |
| 8 | Massachusetts Institute of Technology | 31 |
| 9 | Los Alamos National Laboratory | 31 |
| 10 | Colorado School of Mines | 29 |
| 11 | Pacific Northwest National Laboratory | 28 |
| 12 | Purdue University | 27 |
| 13 | Tokyo Institute of Technology | 25 |
| 14 | Lawrence Livermore National Laboratory | 22 |
| 15 | Faraday Technology, Inc. | 21 |
| 16 | U.S. Naval Research Laboratory | 21 |
| 17 | Tohoku University | 21 |
| 18 | Auburn University | 20 |
| 19 | Kyushu University | 19 |
| 20 | University of Michigan | 18 |
Top 10 affiliations are monopolized by institute in United States. Tokyo Institute of Technology, Rank 13, has the largest counts among institutes outside the US.
Conclusion
Well, I can't reach the topic of natural language processing yet.
But it is interesting to collect basic data and analyze statistically.
In next article, I will start language processing, word embedding by Word2Vec and modification by SCDV.
Original Link: https://dev.to/konkon3249/analysis-of-ecs-236th-meeting-abstracts-1-data-scraping-with-beautifulsoup4-ma8
Dev To
 An online community for sharing and discovering great ideas, having debates, and making friends
An online community for sharing and discovering great ideas, having debates, and making friendsMore About this Source Visit Dev To